Cavity approach for modeling and fitting polymer stretching
Massucci, FA, Pérez Castillo, I, Pérez Vicente, CJ.
Phys. Rev. E
90
,
052708
(2014).
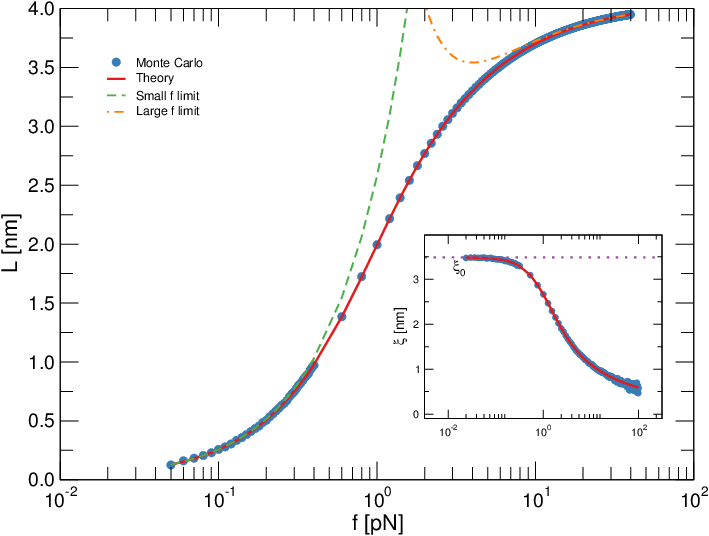
The mechanical properties of molecules are today captured by single molecule manipulation experiments, so that polymer features are tested at a nanometric scale. Yet devising mathematical models to get further insight beyond the commonly studied force-elongation relation is typically hard. Here we draw from techniques developed in the context of disordered systems to solve models for single and double-stranded DNA stretching in the limit of a long polymeric chain. Since we directly derive the marginals for the molecule local orientation, our approach allows us to readily calculate the experimental elongation as well as other observables at wish. As an example, we evaluate the correlation length as a function of the stretching force. Furthermore, we are able to fit successfully our solution to real experimental data. Although the model is admittedly phenomenological, our findings are very sound. For single-stranded DNA our solution yields the correct (monomer) scale and yet, more importantly, the right persistence length of the molecule. In the double-stranded case, our model reproduces the well-known overstretching transition and correctly captures the ratio between native DNA and overstretched DNA. Also in this case the model yields a persistence length in good agreement with consensus, and it gives interesting insights into the bending stiffness of the native and overstretched molecule, respectively.